For a graduate neuronal networks class I took last semester at NYU, I had a project of trying to implement some kind of neuronal network. Given my interest in clustering, I sought to replicate Martí and Rinzel's (2013) feature categorization/clustering network.
Here I show an in-browser demo of their clustering network, as well as a small modification I made to it for anomaly detection. It's kind of fun to play around with (click here to jump directly to it). Most of the parameters in Martí and Rinzel's paper can be modified here to see what effects they have.
The network is an example of a continuous attractor network, which is to say that instead of imagining discrete cells connected to each other, you have an infinite number of cells connected to each other. In some sense, the math becomes easier because you use an integral instead of a sum. However, in this JavaScript demo, I use discrete cells (which can be increased to however many you like, it just makes the simulation slower).
Cells in this network are connected in a ring, and you can imagine each cell as being sensitive to a particular orientation in a line, just like certain visual neurons respond to particular orientations. What this network is going to try to cluster is different orientations that are input into it over time. For example, if the network sees a bunch of nearly flat lines (around $\theta = 0$) over a short period of time, the cells around $\theta = 0$ will begin to maintain a high firing rate. If only a few flat lines are presented though, the cells will forget it and not form a "cluster".
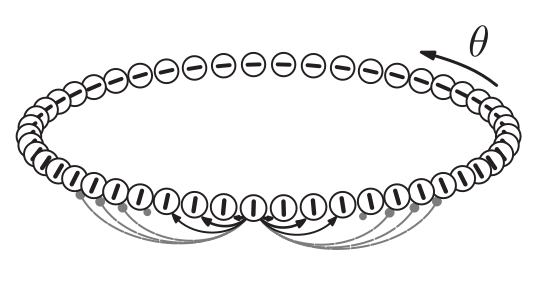
Nearby cells in the ring are connected to each other. With neighboring cells tending to excite each other, slightly further cells tending to inhibit each other, and even further cells having almost no influence. The general shape of the connectivity kernel is what's called a Mexican hat (here approximated with a difference of Gaussians). The general form of this kernel is
\begin{align} J(\theta) &= J_E(\theta) - J_I(\theta)\\ &= j_E \frac{\exp\left(m_E \cos(2\theta)\right)}{I_0(m_E)} - j_I \frac{\exp\left(m_I \cos(2\theta)\right)}{I_0(m_I)} \end{align}
where "$E$" is for excitation and "$I$" inhibition, with $m$ changing the narrowness and $j$ the height of the Mexican hat. "$\theta$" here is the angle difference between neighboring cells. You can mess around with the parameters in the "Kernel" section below to see the effect of different values.
There are two variables that we simulate over time, $s(\theta, t)$, which is the synaptic activation, and $r(\theta, t)$, which represents cell firing. For anomaly detection, I add a third variable $y(\theta, t)$. \begin{align} \tau \frac{\partial}{\partial t}s(\theta, t) &= -s(\theta, t) + r(\theta, t)\\ r(\theta, t) &= \Phi \left[ \frac{1}{\pi} \int_{-\pi/2}^{\pi/2} J(\theta - \theta')s(\theta', t) d\theta' + I(\theta, t)\right]\\ y(\theta, t) &= \Phi(I(\theta, t)) * (1 - s(\theta, t)) \end{align}
Here, $\Phi(x)$ is a sigmoid function for converting a current to a firing rate (between 0 and 1). Its equation is \begin{align} \Phi(x) &= \frac{1}{1 + exp(-\beta[x - x_0])} \end{align} and can be modified in the "Current-to-Rate" section.
The third section, "Excitation", describes how cells respond to stimuli. This basically shows how sensitive cells are to nearby orientations. It has two parameters, $m_s$ and $I_s$ that determine narrowness and the amount of current a cell receives to a nearby orientation. You can modify the inputs to the network by clicking directly on the graph to add more input points. Modifying the "Excitation" section changes how wide and strong each click input is.
Other parameters that can be modified include the length and number of time steps, the number of cells simulated in the network, and the time constant $\tau$.
On the x-axis of the graphs below is time. The y-axis ranges from $-\pi/2$ to $\pi/2$, and represents the orientation of different cells in the network.
This network's fairly interesting. Given the right settings it can detect a number of different clusters of different sizes. With the "anomaly detector" addition, we use the cluster detection to suppress non-novel stimuli. A couple potential problems with this network, though, are that clusters can possibly drift over time and will stay activated indefinitely (which may be problematic for anomaly detection).
Kernel
$jI$: $jE$:
Current-to-Rate
Excitation
Other parameters
Time Step:
Number of time steps:
Delta Theta:
Number of cells:
$\tau$:
Show R:
Anomaly Detector:
 Josh Moller-Mara
Josh Moller-Mara